| Detailed Information for Phage Daredevil |
| Discovery Information |
| Isolation Host | Gordonia terrae 3612 |
| Former names | Aragorn |
| Found By | Elizabeth Crinzi |
| Year Found | 2017 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.439444 N, 79.954167 W Map |
| Discovery Notes | This phage was found in a soil sample collected at Posvar Hall in Pittsburgh, PA on January 8th, 2017 at 3:49 PM EST. The temperature at the time of collection was 16 degrees Fahrenheit, the humidity was 47%, and the weather was cloudy with a 12 mph wind. The day's high temperature 17 degrees Fahrenheit and the low was 8 degrees Fahrenheit. The soil sample was stored in a cool, dark drawer at room temperature for approximately 20 hours before being used in lab. The sample was then directly isolated and incubated for approximately 30 degrees Celsius for three days |
| Naming Notes | I named this phage Daredevil after one of my favorite superheroes and tv shows. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 15, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 680 |
| Genome length (bp) | 90490 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGCAACGCTC |
| GC Content | 64.8% |
| Sequencing Notes | at position 46745 bp approximately 28% of reads contain a C instead of consensus G |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | DL |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
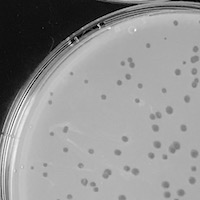
| Plaque Notes | Plaques are about 1.7 mm in diameter. The plaques are clear. |
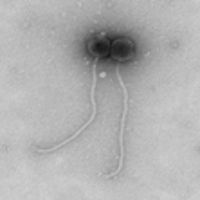
| Morphotype | Siphoviridae |
| Number of Genes | 165 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH590603 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
55 |
| Pitt Freezer Box Grid# |
E7 |
| Available Files |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |

