| Detailed Information for Phage Chilliams |
| Discovery Information |
| Isolation Host | Arthrobacter globiformis B-2979 |
| Found By | Julia Zydanowicz Madision Stevener |
| Year Found | 2024 |
| Location Found | Poughkeepsie, NY America |
| Finding Institution | Marist University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Unknown |
| Isolation Temperature | 30°C |
| GPS Coordinates | 41.72138 N, 73.93435 W Map |
| Discovery Notes | The phage was found during the morning time in dry sand-like soil and had small rocks and sticks in it. |
| Naming Notes | My lab partner and I are on the same cross-country team and we named our phage after our coach; Chuck Williams |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 16, 2024 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 1248 |
| Genome length (bp) | 192062 |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 19328 bp |
| End Determination Notes | There was an area of double coverage about 19-20 kb long apparent when the genome was first assembled, pointing to a likely terminal repeat. The right end of this coverage bump had a clear buildup of read starts, indicating a clear genome terminus. But the left end did not have a clear buildup of read starts, so the left end of the terminal repeat was chosen around where the coverage drops, and is likely not precisely the same in every capsid. |
| GC Content | 53.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | FC |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | San Jacinto Community College |
| Annotation Status | Being Annotated (Expected completion by 5/1/2026) |
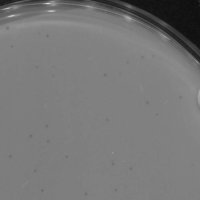
| Plaque Notes | Clear small circular plaques |
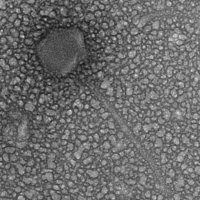
| Morphotype | Siphoviridae |
| Number of Genes | 303 |
| Number of tRNAs | 30 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
5.5 x 10^9 pfu/ml |
| Pitt Freezer Box# |
215 |
| Pitt Freezer Box Grid# |
H1 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |