Arthrobacter phage Piku
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.
| Detailed Information for Phage Piku | |
| Discovery Information | |
| Isolation Host | Arthrobacter globiformis B-2979 |
| Found By | Bisma Khan |
| Year Found | 2022 |
| Location Found | Schaumburg, IL USA |
| Finding Institution | Benedictine University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 22°C |
| GPS Coordinates | 42.034005 N, 88.100313 W Map |
| Discovery Notes | I acquired my soil from a vegetable/fruit garden in Schaumburg, IL. It had rained the previous night which is why it was easy to collect and had a moist texture. I chose this specific location because the garden has been growing plants, vegetables and fruits such as flowers, spearmint leaves, coriander, tomatoes and peppers for years. This location has been topped with fertilizer, compost and soil multiple times which is why it is a prime spot for bacterial growth and an ideal place to extract a phage. In order to collect my sample I used gloves to avoid contaminating the environmental sample and a small garden shovel. I made sure to keep the sample free from any solid/hard pieces, wood chips, and weeds in order to ensure the filtration process goes smoothly. I acquired exactly 5 grams of the spoil sample. The soils physical characteristics were dark and moist, almost a clay like structure. |
| Naming Notes | I chose the name piku becuase it was the name of a pet cat I used to have. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Feb 20, 2023 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 6511 |
| Genome length (bp) | 15675 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 15 bases |
| Overhang Sequence | CCACGGTCCCCGTCC |
| GC Content | 64.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | FE |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
Click to View |
| Annotating Institution | Benedictine University |
| Annotation Status | In GenBank |
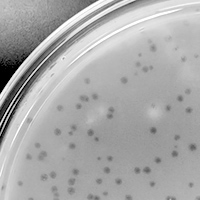
| Plaque Notes | plaque diameter: 1mm plaque radius: 0.5 Page piku formed clear lytic plaques with the diameter of 1mm and a radius of 0.5mm. Since my radius was smaller, a higher PFU/ml was required in order to create webbed plates. The temperature that the phage grew the best at was 22C. This tells us that the phage amplifies better at lower temperatures rather than high. This phage belongs to the Podoviridae family becuase it's characteristics include short, noncontractile tails. |
| Morphotype | Siphoviridae |
| Number of Genes | 22 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewPiku_1 Piku_2 Piku_3 Piku_4 Piku_5 Piku_6 Piku_7 Piku_8 Piku_9 Piku_10 Piku_11 Piku_12 Piku_13 Piku_14 Piku_15 Piku_16 Piku_17 Piku_18 Piku_19 Piku_20 Piku_21 Piku_22 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PQ362670 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 217 |
| Pitt Freezer Box Grid# | G4 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |