| Detailed Information for Phage RexFury |
| Discovery Information |
| Isolation Host | Rhodococcus erythropolis RIA 643 |
| Found By | Brent Cunningham and Conner Satterlund |
| Year Found | 2014 |
| Location Found | River Falls, WI USA |
| Finding Institution | University of Wisconsin-River Falls |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Unknown |
| Isolation Temperature | 30°C |
| GPS Coordinates | 44.840008 N, 92.648331 W Map |
| Naming Notes | soil sample was initially called RF for Rhodo field soil, extended that to a name that seemed to fit our phage's ability to quickly devour bacteria |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 26, 2015 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 150 |
| Genome length (bp) | 46627 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGCCGTGAT |
| GC Content | 58.6% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | CA |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
| Plaque Notes | large, clear plaques |
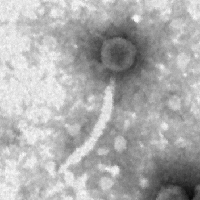
| Morphotype | Siphoviridae |
| Number of Genes | 65 |
| Number of tRNAs | 3 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MF324904 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
12 |
| Pitt Freezer Box Grid# |
H3 |
| Available Files |
| EM Picture | Download |
| GenBank File for Phamerator | Download |
