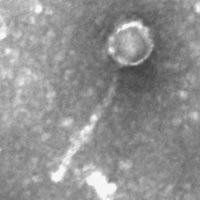
| Detailed Information for Phage Naiad | |
| Discovery Information | |
| Isolation Host | Rhodococcus erythropolis RIA 643 |
| Found By | Bradley Koch and Brittany Lubich |
| Year Found | 2014 |
| Location Found | Hudson, WI USA |
| Finding Institution | University of Wisconsin-River Falls |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 44.979117 N, 92.759906 W Map |
| Discovery Notes | soil from east bank of St Croix River, near kayak launch site, next to small stream running from the river to a walking path We think Naiad is a clear plaque mutant of StCroix. They have 2 nt differences and one is in the repressor gene, which change an Arg to a Leu near the HTH region |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 21, 2015 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina Sequencing |
| Approximate Shotgun Coverage | 3861 |
| Genome length (bp) | 46619 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGCCGTGAT |
| End Determination Notes | Clear read buildups in Illumina sequencing data. |
| GC Content | 58.6% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CA |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
Click to ViewAlatin Alpacados AngryOrchard AppleCloud Belenaria BobbyDazzler Bonanza Bradshaw Bryce CosmicSans Dinger Erik Espica Gollum Harlequin Hiro Jester Krishelle Lillie Naiad Nancinator Natosaleda Partridge PhailMary Phrankenstein Rasputin RER2 RexFury RGL3 Rhodalysa Shuman StCroix Swann Takoda TWAMP UhSalsa Yogi Yoncess |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
| Plaque Notes | large, clear plaques, 1-2 mm diameter, appearing at 24 hr. Separated from turbid plaques in same sample (turbid plaques isolated as StCroix) |
| Morphotype | Siphoviridae |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewNaiad_1 Naiad_2 Naiad_3 Naiad_7 Naiad_8 Naiad_9 Naiad_10 Naiad_11 Naiad_12 Naiad_13 Naiad_14 Naiad_15 Naiad_16 Naiad_17 Naiad_18 Naiad_19 Naiad_20 Naiad_21 Naiad_22 Naiad_23 Naiad_24 Naiad_25 Naiad_26 Naiad_27 Naiad_28 Naiad_29 Naiad_30 Naiad_31 Naiad_32 Naiad_33 Naiad_34 Naiad_35 Naiad_36 Naiad_37 Naiad_38 Naiad_39 Naiad_40 Naiad_41 Naiad_42 Naiad_43 Naiad_44 Naiad_45 Naiad_46 Naiad_47 Naiad_48 Naiad_49 Naiad_50 Naiad_51 Naiad_52 Naiad_53 Naiad_54 Naiad_55 Naiad_56 Naiad_57 Naiad_58 Naiad_59 Naiad_60 Naiad_61 Naiad_62 Naiad_63 Naiad_64 Naiad_65 Naiad_66 Naiad_67 Naiad_68 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MF324901 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 1.16 x 10^10 |
| Date of SEA Lysate Titering | Oct 30, 2014 |
| Pitt Freezer Box# | 12 |
| Pitt Freezer Box Grid# | H2 |
| Available Files | |
| EM Picture | Download |
| GenBank File for Phamerator | Download |