| Detailed Information for Phage RikSengupta |
| Discovery Information |
| Isolation Host | Microbacterium foliorum NRRL B-24224 |
| Found By | Abhirup Das |
| Year Found | 2022 |
| Location Found | Amherst, MA United States of America |
| Finding Institution | Illinois Wesleyan University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 42.37595 N, 72.504425 W Map |
| Discovery Notes | The soil sample was taken from underneath a rock, beneath a shrub, in a backyard covered in dense organic material. The soil was covered with semi-decomposed leaves, and twigs. There were colonies of ants nearby. The area also had several other insects, including crickets, and spiders. The soil was dry and dark brown in color. The ambient temperature was approximately 29°C. |
| Naming Notes | The name of the phage is after a friend who helped me document the collection of the soil sample from which I was able to isolate the phage. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 4, 2023 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 723 |
| Genome length (bp) | 57187 |
| Character of genome ends | Circularly Permuted |
| GC Content | 62.7% |
| Sequencing Notes | Around 15% of the reads from this sample were from a novel Cluster EE phage which was likely a contaminant. |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | EF |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Illinois Wesleyan University |
| Annotation Status | In GenBank |
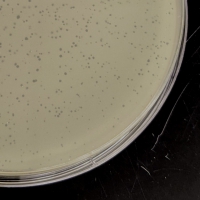
| Plaque Notes | Plaques are almost uniformly tiny, and always clear. |
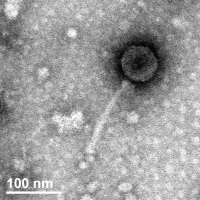
| Morphotype | Siphoviridae |
| Number of Genes | 87 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | OQ995427 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
152 |
| Pitt Freezer Box Grid# |
F2 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |