| Detailed Information for Phage Nova |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Joe McMaster (Nova) |
| Year Found | 2006 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | Unknown |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.446526 N, 79.954479 W Map |
| Naming Notes | A representative from Nova came to check out our program. though we didn't make it to a TV show, we were able to isolate a phage from the sample that Joe dug from the dirt. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Sep 20, 2011 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | 454 |
| Sequencer Used | 454 Genome Sequencer FLX |
| Approximate Shotgun Coverage | 105 |
| Genome length (bp) | 65108 |
| Character of genome ends | Circularly Permuted |
| GC Content | 59.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | D |
| Subcluster | D1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
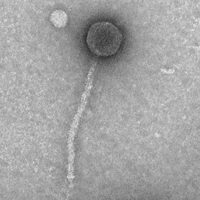
| Morphotype | Siphoviridae |
| Number of Genes | 88 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | JN699014 |
| Refseq Number | None yet |
| Published in Paper | Pope, Bowman, Russell, et al 2015, eLIFE: Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
2 |
| Pitt Freezer Box Grid# |
E6 |
| Available Files |
| Plaque Picture | Download |
| Virtual Digest Picture | Download |
| EM Picture | Download |
| Final DNAMaster File | Download |