| Detailed Information for Phage Wayne |
| Discovery Information |
| Isolation Host | Arthrobacter sp. ATCC 21022 |
| Found By | Deborah Jacobs-Sera |
| Year Found | 2012 |
| Location Found | USA |
| Finding Institution | University of Pittsburgh |
| Program | Phage Hunters Integrating Research and Education |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.326528 N, 79.677959 W Map |
| Discovery Notes | Sample was obtained in a front yard island bed of rhododendron and ivy below a Canada Red cherry tree. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Mar 28, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 3498 |
| Genome length (bp) | 44371 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 13 bases |
| Overhang Sequence | GGTAACCGTGATA |
| GC Content | 61.1% |
| Sequencing Notes | Originally seeunced with Ion Torent. Re-sequenced with Illumina (3/2017). The changes are: 16001 G substitution for A. 17906 T substitution for G, and 19091 C for G, and 19109 C substitution for A. C insertion at 35247. |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | AK |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
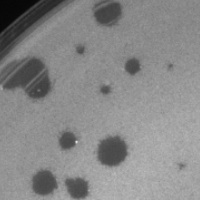
| Plaque Notes | Plaque morphology is clear and round, ranging in diameter from 1 to 3 mm. |
| Morphotype | Siphoviridae |
| Number of Genes | 63 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | KU160672 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
5 |
| Pitt Freezer Box Grid# |
A2 |
| Available Files |
| Plaque Picture | Download |
| Virtual Digest Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |
