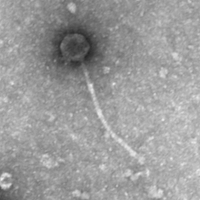
| Detailed Information for Phage Cursive |
| Discovery Information |
| Isolation Host | Streptomyces lividans JI 1326 |
| Found By | Christopher Festin and Katie Rector |
| Year Found | 2022 |
| Location Found | St. Louis, MO United States of America |
| Finding Institution | Washington University in St. Louis |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 38.432708 N, 90.446399 W Map |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Feb 16, 2023 |
| Sequencing Facility | The McDonnell Genome Institute at Washington University |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 152 |
| Genome length (bp) | 131901 |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 9892 bp |
| GC Content | 49.8% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | BE |
| Subcluster | BE1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Washington University in St. Louis |
| Annotation Status | In GenBank |
| Number of Genes | 231 |
| Number of tRNAs | 43 |
| Number of tmRNAs | 1 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PP405139 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |
