| Detailed Information for Phage Procrass1 | |
| Discovery Information | |
| Isolation Host | Propionibacterium acnes ATCC 6919 |
| Former names | PAPB3 |
| Found By | Viviana Petru |
| Year Found | 2011 |
| Location Found | Los Angeles, CA USA |
| Finding Institution | University of California, Los Angeles |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 37°C |
| GPS Coordinates | 34.065 N, 118.4425 W Map |
| Discovery Notes | Phage isolation performed by Rania Abolhosn, Mandip Kang, Trung Luu, and Viviana Petru (UCLA). DNA preparation performed by Krisanavane Reddi (UCLA). Assembly performed by William Villella (UCLA) and confirmed by Charles Bowman (University of Pittsburgh). Genome annotation performed by David Gray, Taryn McLaughlin, Rachel Stafford-Lewis, Aprajita Yadav, and Kylie Wilson (UCLA). Quality control of annotation results performed by Jordan Moberg Parker and Dallas Mould (UCLA). |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 26, 2015 |
| Sequencing Facility | The UCLA Genotyping and Sequencing Core |
| Shotgun Sequencing Method | 454 |
| Sequencer Used | 454 Genome Sequencer FLX |
| Approximate Shotgun Coverage | 885 |
| Genome length (bp) | 29347 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | TCGTACGGCTT |
| GC Content | 54.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | BU |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
Click to ViewAquarius ATCC29399BC ATCC29399BT Attacne BruceLethal Cota DrParker Enochoraptor Enoki Keiki Kubed Lauchelly Leviosa LilBandit MEAK Moyashi MrAK Ouroboros P1.1 P100.1 P100A P100D P101A P104A P104B P105 P106A P106C P106I P106L P106M P107A P107C P108C P14.4 P9.1 PA6 PAD20 PAS50 Pirate Procrass1 QueenBey Rileysaurus SKKY Solid Stormborn Supernova Wizzo |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
| Plaque Notes | small, clear, circular plaques |
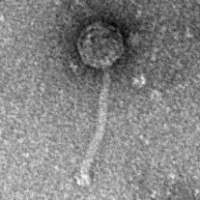
| Morphotype | Siphoviridae |
| Number of Genes | 45 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewProcrass1_1 Procrass1_2 Procrass1_3 Procrass1_4 Procrass1_5 Procrass1_6 Procrass1_7 Procrass1_8 Procrass1_9 Procrass1_10 Procrass1_11 Procrass1_12 Procrass1_13 Procrass1_14 Procrass1_15 Procrass1_16 Procrass1_17 Procrass1_18 Procrass1_19 Procrass1_20 Procrass1_21 Procrass1_22 Procrass1_23 Procrass1_24 Procrass1_25 Procrass1_26 Procrass1_27 Procrass1_28 Procrass1_29 Procrass1_30 Procrass1_31 Procrass1_32 Procrass1_33 Procrass1_34 Procrass1_35 Procrass1_36 Procrass1_37 Procrass1_38 Procrass1_39 Procrass1_40 Procrass1_41 Procrass1_42 Procrass1_43 Procrass1_44 Procrass1_45 |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | KR337644 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Designator | 2011UCLAbruinVP |
| SEA Lysate Titer | 5.2x10^9 |
| Date of SEA Lysate Titering | Nov 21, 2011 |
| Pitt Freezer Box# | 28 |
| Pitt Freezer Box Grid# | D3 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| Final DNAMaster File | Download |