| Detailed Information for Phage Prospero | |
| Discovery Information | |
| Isolation Host | Arthrobacter sp. ATCC 21022 |
| Found By | Joseph Quinlan, Jr. |
| Year Found | 2015 |
| Location Found | Vineland, NJ USA |
| Finding Institution | Saint Joseph's University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 23°C |
| GPS Coordinates | 39.486400 N, 75.026000 W Map |
| Discovery Notes | The phage was derived from top soil that was collected in a back yard in Vineland, on a humid night, near a stone driveway. The soil was slightly moist due to condensation. |
| Naming Notes | The name Prospero is derived from two topics. First, it was chosen due to the phage's high, or prosperous, success rate in lab procedures. Secondly, the name was found in the play the Tempest by the great playwright William Shakespeare. Prospero is an older Italian name that roughly translates to "prosperous one". |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 13, 2016 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 2069 |
| Genome length (bp) | 15556 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CCCGCGCCACC |
| GC Content | 60.1% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | AN |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Lysogeny Notes | Prospero infected its bacterial host rather rapidly, often taking only 24 hours or less to achieve the desired results in the infection procedure. |
| Other Cluster Members |
Click to ViewArby Arielagos Azathoth Blair CGermain Chestnut Copper Courtney3 Decurro Dewayne ElectricPheel Elkhorn Eyoseyas FirePit Guntur Hunnie Inspire2 Jessica Kels KylieMac Laila Link Lore LouisXIV Maggie Mariposa Massimo Moloch Muttlie Prospero Ronnie Sandman Saphira SerialPhiller Seume Sourignavong StewieGriff Stratus StrawberryLem Swenson Taj14 TinoCrisci Toulouse TymAbreu Vinye Ximeno Yank |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
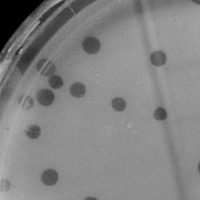
| Plaque Notes | The plaques of Prospero ranged from 3.0 to 1.0 mm in diameter, all of which were colorless and circular. |
| Morphotype | Siphoviridae |
| Number of Genes | 26 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewProspero_1 Prospero_2 Prospero_3 Prospero_4 Prospero_5 Prospero_6 Prospero_7 Prospero_8 Prospero_9 Prospero_10 Prospero_11 Prospero_12 Prospero_13 Prospero_14 Prospero_15 Prospero_16 Prospero_17 Prospero_18 Prospero_19 Prospero_20 Prospero_21 Prospero_22 Prospero_23 Prospero_24 Prospero_25 Prospero_26 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | KX610765 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 26 |
| Pitt Freezer Box Grid# | G5 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |