| Detailed Information for Phage Pharky |
| Discovery Information |
| Isolation Host | Microbacterium foliorum NRRL B-24224 |
| Found By | Aleda Jomy |
| Year Found | 2019 |
| Location Found | Philadelphia, PA United States of America |
| Finding Institution | Drexel University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 25°C |
| GPS Coordinates | 39.961111 N, 75.188056 W Map |
| Discovery Notes | The sample was collected from an open grassy area with trees at Drexel Park, on the Drexel University campus. The park soil was slightly damp. |
| Naming Notes | I found the phage in the park, so my partner and I decided to replace the p in park with a ph, and then we added the -y to make it cute. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 4, 2019 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 192 |
| Genome length (bp) | 54442 |
| Character of genome ends | Circularly Permuted |
| GC Content | 59.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | EK |
| Subcluster | EK2 |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
|
| Annotating Institution | Drexel University |
| Annotation Status | In GenBank |
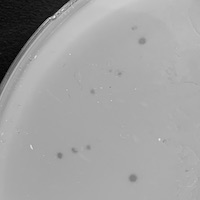
| Plaque Notes | The plaque assay had around 15-20 plaques of varying morphologies. They all seem to be clear plaques, but of varying sizes. The smaller plaques were 0.8mm and the larger ones were around 1 mm. |
| Number of Genes | 55 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MT310891 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
105 |
| Pitt Freezer Box Grid# |
H2 |
| Available Files |
| Plaque Picture | Download |
| GenBank File for Phamerator | Download |
