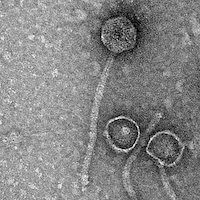
Mycobacterium phage Phreak
Add or modify phage thumbnail images to appear at the top of this page.
Know something about this phage that we don't? Modify its data.