| Detailed Information for Phage Tyrumbra |
| Discovery Information |
| Isolation Host | Microbacterium paraoxydans NRRL B-14843 |
| Found By | Christopher Jentzsch |
| Year Found | 2018 |
| Location Found | Hudson, WI USA |
| Finding Institution | University of Wisconsin-River Falls |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 28°C |
| GPS Coordinates | 44.973333 N, 92.731667 W Map |
| Discovery Notes | collected on sept 9 at 4:54pm in a garden in Hudson WI.The humidity was 53% on a cloudy day. The soil was watered every 3 days. |
| Naming Notes | The name came from my D&D campaign |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Feb 18, 2019 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina Sequencing |
| Approximate Shotgun Coverage | 205 |
| Genome length (bp) | 53975 |
| Character of genome ends | Circularly Permuted |
| GC Content | 68.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | EC |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | University of Wisconsin-River Falls |
| Annotation Status | In GenBank |
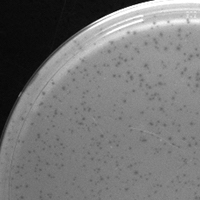
| Plaque Notes | small uniform evenly distributed plaques, lacking halos or cloudyness. |
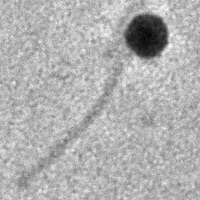
| Morphotype | Siphoviridae |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MN175603 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
88 |
| Pitt Freezer Box Grid# |
B2 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |