| Detailed Information for Phage BigAnti | |
| Discovery Information | |
| Isolation Host | Corynebacterium glutamicum MB001 |
| Found By | Binyam Assefa |
| Year Found | 2025 |
| Location Found | Ipswich, MA United States |
| Finding Institution | Harvard University |
| Program | Community Phages |
| From enriched soil sample? | Unknown |
| Isolation Temperature | Not entered |
| GPS Coordinates | 42.6472 N, 70.84894 W Map |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 9, 2026 |
| Sequencing Facility | Harvard University |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 740 |
| Genome length (bp) | 53420 |
| Character of genome ends | Circularly Permuted |
| End Determination Notes | This was the first of its cluster, and was actually sequenced twice, once at Harvard and once at Pitt. Resulting assemblies were checked for any obvious buildup of read starts or coverage anomalies, but none were found, implying that this phage packages its DNA in a headful, circularly permuted way. |
| GC Content | 57.9% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | Singleton |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to ViewArV2 Attoomi Ball Barebow BigAnti Cantare ChewyVIII Chymera DillyDally DocB7 DS6A E3 Elener EmiRose ERHickory Eyre Finch Finkle Footloose FuzzBuster GAL1 Gilgamesh GMA1 GMA4 Gudmit Ibantik Jace JeanGrey Juicebox Kithara Kromp Kumao LilSpotty LuckyBarnes Lukepolites Magritte Min1 MooMoo mu16 OnionKnight P3MA PensacolaC28 phiAsp2 phiSAV Phroglets Pine5 Ponzi pZL12 REQ2 REQ3 Reynauld Root RRH1 Samy Shambre1 Shocker Shrew Sleepyhead Spartoi Sunfish Tailonex TinyDot TJE1 Tonitrus TPA2 TPA4 Trogglehumper TurkishDelight Whack Wollypog Yappy |
| Annotating Institution | Harvard University |
| Annotation Status | Being Annotated (Expected completion by 5/1/2026) |
| Number of Genes | 68 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewBigAnti_1 BigAnti_2 BigAnti_3 BigAnti_4 BigAnti_5 BigAnti_6 BigAnti_7 BigAnti_8 BigAnti_9 BigAnti_10 BigAnti_11 BigAnti_12 BigAnti_13 BigAnti_14 BigAnti_15 BigAnti_16 BigAnti_17 BigAnti_18 BigAnti_19 BigAnti_20 BigAnti_21 BigAnti_22 BigAnti_23 BigAnti_24 BigAnti_25 BigAnti_26 BigAnti_27 BigAnti_28 BigAnti_29 BigAnti_30 BigAnti_31 BigAnti_32 BigAnti_33 BigAnti_34 BigAnti_35 BigAnti_36 BigAnti_37 BigAnti_38 BigAnti_39 BigAnti_40 BigAnti_41 BigAnti_42 BigAnti_43 BigAnti_44 BigAnti_45 BigAnti_46 BigAnti_47 BigAnti_48 BigAnti_49 BigAnti_50 BigAnti_51 BigAnti_52 BigAnti_53 BigAnti_54 BigAnti_55 BigAnti_56 BigAnti_57 BigAnti_58 BigAnti_59 BigAnti_60 BigAnti_61 BigAnti_62 BigAnti_63 BigAnti_64 BigAnti_65 BigAnti_66 BigAnti_67 BigAnti_68 |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Not in Pitt Archives |
| Available Files | |
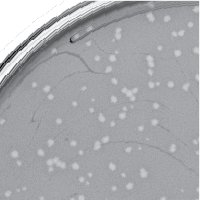
| Plaque Picture | Download |
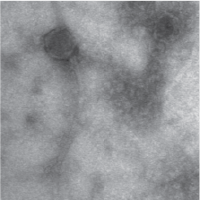
| EM Picture | Download |