| Detailed Information for Phage LilSpotty | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Rinah Kim |
| Year Found | 2018 |
| Location Found | La Crescenta, CA USA |
| Finding Institution | University of California, Los Angeles |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 35°C |
| GPS Coordinates | 34.223687 N, 118.255645 W Map |
| Discovery Notes | Phage LilSpotty was isolated from moderately moist, dark soil on 4/8/2018 by Rinah Kim at a depth of 4 inches. Air temp at time of collection was 14˚C. The collection site distant from buildings; moderately low foot traffic; approximately 50 m from the major road and cement; no water in close proximity; many trees (more than 15) present within the range of 10 m (the site was located in a local park); slightly sloped. Phage LilSpotty was isolated by standard enrichment protocol at 35˚C. High-Titer Lysate [Midi-Lysate] (Archive) Titer: 1.6 * 10^10 Date of High-Titer Lysate Titer: 5/23/18 Characterization was performed by Team We're Goin' Viral (Christian Torres, Kelly Duong, Micah Chong, Chris Lee) and Team KARL (Karina Haleblian, Alexis Ballew, Lillian Lai, Rinah Kim). MIMG 103AL-S18 Instructor: Dr. Amanda Freise Teaching Assistant: Samuel Wu Undergraduate Assistants: Kalin Patel & Mariam Khatchatrian Laboratory Support: Dr. Krisanavane Reddi and Dr. Jordan Moberg Parker |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Aug 3, 2018 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 2612 |
| Genome length (bp) | 49798 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGCCGGCAT |
| End Determination Notes | Ends were fairly apparent from the buildup of reads flanking an overhang, though the right end of the genome had fewer obvious reads supporting the end call. |
| GC Content | 64.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | Singleton |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to ViewArV2 Attoomi Ball Barebow Cantare ChewyVIII Chymera CyranoPS DillyDally DocB7 DS6A E3 Elener EmiRose Eyre Finch Finkle Footloose FuzzBuster GAL1 Gilgamesh GMA1 GMA4 Gudmit Ibantik Jace JeanGrey Juicebox Kithara Kromp Kumao LilSpotty LuckyBarnes Lukepolites Magritte Min1 MooMoo mu16 OnionKnight P3MA PensacolaC28 phiAsp2 phiSAV Phroglets Pine5 Ponzi pZL12 REQ2 REQ3 Reynauld RomansRevenge Root RRH1 Samy Shambre1 Shocker Shrew Sleepyhead Spartoi Sunfish TinyDot TJE1 Tonitrus TPA2 TPA4 Trogglehumper TurkishDelight ValentiniPuff Whack Wollypog Yappy |
| Annotating Institution | University of California, Los Angeles |
| Annotation Status | In GenBank |
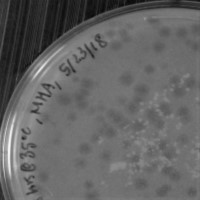
| Plaque Notes | Plaque Morphology: From the enrichment isolation plates, most of the plaque morphology is tiny and clear but there were some plaques that were turbid. Average diameter in mm: 2.0 mm |
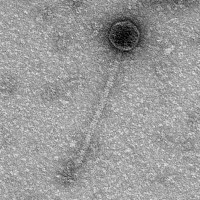
| Morphotype | Siphoviridae |
| Number of Genes | 85 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewLilSpotty_1 LilSpotty_2 LilSpotty_3 LilSpotty_4 LilSpotty_5 LilSpotty_6 LilSpotty_7 LilSpotty_8 LilSpotty_9 LilSpotty_10 LilSpotty_11 LilSpotty_12 LilSpotty_13 LilSpotty_14 LilSpotty_15 LilSpotty_16 LilSpotty_17 LilSpotty_18 LilSpotty_19 LilSpotty_20 LilSpotty_21 LilSpotty_22 LilSpotty_23 LilSpotty_24 LilSpotty_25 LilSpotty_26 LilSpotty_27 LilSpotty_28 LilSpotty_29 LilSpotty_30 LilSpotty_31 LilSpotty_32 LilSpotty_33 LilSpotty_34 LilSpotty_35 LilSpotty_36 LilSpotty_37 LilSpotty_38 LilSpotty_39 LilSpotty_40 LilSpotty_41 LilSpotty_42 LilSpotty_43 LilSpotty_44 LilSpotty_45 LilSpotty_46 LilSpotty_47 LilSpotty_48 LilSpotty_49 LilSpotty_50 LilSpotty_51 LilSpotty_52 LilSpotty_53 LilSpotty_54 LilSpotty_55 LilSpotty_56 LilSpotty_57 LilSpotty_58 LilSpotty_59 LilSpotty_60 LilSpotty_61 LilSpotty_62 LilSpotty_63 LilSpotty_64 LilSpotty_65 LilSpotty_66 LilSpotty_67 LilSpotty_68 LilSpotty_69 LilSpotty_70 LilSpotty_71 LilSpotty_72 LilSpotty_73 LilSpotty_74 LilSpotty_75 LilSpotty_76 LilSpotty_77 LilSpotty_78 LilSpotty_79 LilSpotty_80 LilSpotty_81 LilSpotty_82 LilSpotty_83 LilSpotty_84 LilSpotty_85 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MK977707 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 78 |
| Pitt Freezer Box Grid# | C5 |
| Available Files | |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |