| Detailed Information for Phage RomansRevenge | |
| Discovery Information | |
| Isolation Host | Arthrobacter sulfureus ATCC 19098 |
| Found By | Kylie Tseng |
| Year Found | 2023 |
| Location Found | Los Angeles, CA US |
| Finding Institution | University of California, Los Angeles |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 34.06202 N, 118.29652 W Map |
| Discovery Notes | Isolation by Dominic Garza and Michelle Zorawik. An enriched culture was prepared by combining 15 mL of soil with 25 mL of PYCA broth. The bacterial strains of A. sp., A. globiformis, A. sulfureus and A. atrocyaneus were then added in equal concentrations. The enriched culture was incubated at 30℃ for 48 hours with shaking. After filtering with a 0.22 µm filter, the sample was plated on PYCA plates with 2X PYCa soft agar using the standard pour plate method. Characterization by Team PhageFright. Milena Deal, Nicole Critzer, Thomas Kretschmer, Griffin Gowdy, Audia Hughes AL Quarter and Lab section: W23-1B Instructor: Amanda Freise Teaching Assistant: Dom Garza Undergraduate Assistant(s): Melvin and Sophia |
| Naming Notes | The name is a direct reference to a song on Nicki Minaj’s debut studio album titled Pink Friday. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jul 19, 2023 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 293 |
| Genome length (bp) | 62027 |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 1042 bp |
| End Determination Notes | As this was the first phage of its (close) kind, we looked closely at the ends. There was a clear region of about 1 kb that had ~2x the coverage level of the rest of the genome. This implies a direct terminal repeat. The right end of this terminal repeat was well-defined in the sequencing reads, with many read starts appearing at a particular base. The left end was less well-defined, however, and so we chose a base near the left end of the double-coverage region that had slightly more read starts than the surrounding area. |
| GC Content | 60.6% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | FT |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to View |
| Annotating Institution | University of California, Los Angeles |
| Annotation Status | In GenBank |
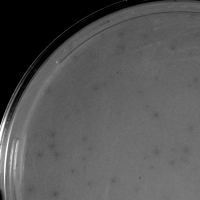
| Plaque Notes | Very turbid with pinpoint clear centers. 1 mm |
| Number of Genes | 93 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewRomansRevenge_1 RomansRevenge_2 RomansRevenge_3 RomansRevenge_4 RomansRevenge_5 RomansRevenge_6 RomansRevenge_7 RomansRevenge_8 RomansRevenge_9 RomansRevenge_10 RomansRevenge_11 RomansRevenge_12 RomansRevenge_13 RomansRevenge_14 RomansRevenge_15 RomansRevenge_16 RomansRevenge_17 RomansRevenge_18 RomansRevenge_19 RomansRevenge_20 RomansRevenge_21 RomansRevenge_22 RomansRevenge_23 RomansRevenge_24 RomansRevenge_25 RomansRevenge_26 RomansRevenge_27 RomansRevenge_28 RomansRevenge_29 RomansRevenge_30 RomansRevenge_31 RomansRevenge_32 RomansRevenge_33 RomansRevenge_34 RomansRevenge_35 RomansRevenge_36 RomansRevenge_37 RomansRevenge_38 RomansRevenge_39 RomansRevenge_40 RomansRevenge_41 RomansRevenge_42 RomansRevenge_43 RomansRevenge_44 RomansRevenge_45 RomansRevenge_46 RomansRevenge_47 RomansRevenge_48 RomansRevenge_49 RomansRevenge_50 RomansRevenge_51 RomansRevenge_52 RomansRevenge_53 RomansRevenge_54 RomansRevenge_55 RomansRevenge_56 RomansRevenge_57 RomansRevenge_58 RomansRevenge_59 RomansRevenge_60 RomansRevenge_61 RomansRevenge_62 RomansRevenge_63 RomansRevenge_64 RomansRevenge_65 RomansRevenge_66 RomansRevenge_67 RomansRevenge_68 RomansRevenge_69 RomansRevenge_70 RomansRevenge_71 RomansRevenge_72 RomansRevenge_73 RomansRevenge_74 RomansRevenge_75 RomansRevenge_76 RomansRevenge_77 RomansRevenge_78 RomansRevenge_79 RomansRevenge_80 RomansRevenge_81 RomansRevenge_82 RomansRevenge_83 RomansRevenge_84 RomansRevenge_85 RomansRevenge_86 RomansRevenge_87 RomansRevenge_88 RomansRevenge_89 RomansRevenge_90 RomansRevenge_91 RomansRevenge_92 RomansRevenge_93 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PV915819 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 184 |
| Pitt Freezer Box Grid# | E9 |
| Available Files | |
| Plaque Picture | Download |
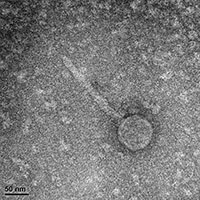
| EM Picture | Download |
| GenBank File for Phamerator | Download |