| Detailed Information for Phage Attoomi | |
| Discovery Information | |
| Isolation Host | Streptomyces azureus NRRL B-2655 |
| Found By | Ahmad Sulaiman |
| Year Found | 2016 |
| Location Found | New Braunfels, TX USA |
| Finding Institution | University of North Texas |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 24°C |
| GPS Coordinates | 29.711028 N, 98.133833 W Map |
| Discovery Notes | The soil sample was collected from Landa Park in New Braunfels, TX. The temperatures was 24°C, and the soil was wet. The sample was taken 2 cm under surface. |
| Naming Notes | It's a nickname for my wife that she likes the most. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jun 27, 2017 |
| Sequencing Facility | University of North Texas |
| Shotgun Sequencing Method | Illumina |
| Genome length (bp) | 41872 |
| Character of genome ends | Unknown |
| Overhang Length | 11 bases |
| Overhang Sequence | CGAAACGTGAT |
| GC Content | 69.7% |
| Sequencing Notes | There was a bizarre coverage pattern for this genome. More than 17,000 of the ~70,000 reads in the assembly began at exactly base 1. Usually, we see a buildup of reads at base 1 of defined-end genomes, but this was orders of magnitude higher than normal. This resulted in coverage levels of ~18000 for the first couple kb, dropping to about 500 after ~3 kb, and level out around 100 thereafter. We're not sure what caused this, but it does appear from other evidence that this genome has defined ends and a 3' overhang, so it is reported as such. |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | Singleton |
| Subcluster | -- |
| Cluster Life Cycle | Unknown |
| Other Cluster Members |
Click to ViewArV2 Attoomi Ball Barebow BigAnti Cantare ChewyVIII Chymera DillyDally DocB7 DS6A E3 Elener EmiRose ERHickory Eyre Finch Finkle Footloose FuzzBuster GAL1 Gilgamesh GMA1 GMA4 Gudmit Ibantik Jace JeanGrey Juicebox Kithara Kromp Kumao LilSpotty LuckyBarnes Lukepolites Magritte Min1 MooMoo mu16 OnionKnight P3MA PensacolaC28 phiAsp2 phiSAV Phroglets Pine5 Ponzi pZL12 REQ2 REQ3 Reynauld Root RRH1 Samy Shambre1 Shocker Shrew Sleepyhead Spartoi Sunfish Tailonex TinyDot TJE1 Tonitrus TPA2 TPA4 Trogglehumper TurkishDelight Whack Wollypog Yappy |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
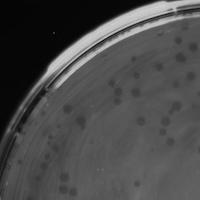
| Plaque Notes | The diameter is 1.5 mm . It makes clear zone in spot test |
| Number of Genes | 53 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewAttoomi_1 Attoomi_2 Attoomi_3 Attoomi_4 Attoomi_5 Attoomi_6 Attoomi_7 Attoomi_8 Attoomi_9 Attoomi_10 Attoomi_11 Attoomi_12 Attoomi_13 Attoomi_14 Attoomi_15 Attoomi_16 Attoomi_17 Attoomi_18 Attoomi_19 Attoomi_20 Attoomi_21 Attoomi_22 Attoomi_23 Attoomi_24 Attoomi_25 Attoomi_26 Attoomi_27 Attoomi_28 Attoomi_29 Attoomi_30 Attoomi_31 Attoomi_32 Attoomi_33 Attoomi_34 Attoomi_35 Attoomi_36 Attoomi_37 Attoomi_38 Attoomi_39 Attoomi_40 Attoomi_41 Attoomi_42 Attoomi_43 Attoomi_44 Attoomi_45 Attoomi_46 Attoomi_47 Attoomi_48 Attoomi_49 Attoomi_50 Attoomi_51 Attoomi_52 Attoomi_53 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MG593801 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 4x10^8 pfu/mL |
| Pitt Freezer Box# | 36 |
| Pitt Freezer Box Grid# | C12 |
| Available Files | |
| Plaque Picture | Download |
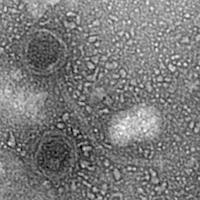
| EM Picture | Download |
| GenBank File for Phamerator | Download |