| Detailed Information for Phage William | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Parampreet Deol |
| Year Found | 2016 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.443383 N, 79.955009 W Map |
| Discovery Notes | The soil was collected from a flower bed outside a clear, sunny day. The soil was slightly moist, and I dug about 3 inches into the ground to collect it. Afterwards, it was stored in a refrigerator for about 1 day before being processed in lab. |
| Naming Notes | The name for this phage comes from the location it was found. It was found outside of the William Pitt Union at the University of Pittsburgh. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Feb 16, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 456 |
| Genome length (bp) | 50678 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | TCGCCGGTGA |
| GC Content | 66.5% |
| Sequencing Notes | There were two SNPs in this sample. At position 23844, ~40% of the reads showed a G instead of the consensus A. At position 24218, ~35% of the reads showed a G instead of the consensus A. |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CV |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
Click to ViewArchis Azula Barco Begonia BlingBling Blino Blueberry CaptainKirk2 CarolAnn CheeseSpider Delian Fairfaxidum Fenry Frokostdame Gambino Guacamole Hitter Jalammah JasperJr JuJu KappaFarmDelta Lysidious Malachai Melba Mellie MintFen MissRona Murp Obliviate PCoral7 Petra PhrostedPhlake Pipp PrincePatrick Samba Toast UmaThurman Utz Walrus William Wisp Wocket Zarbodnamra ZiggyZoo Zilla |
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
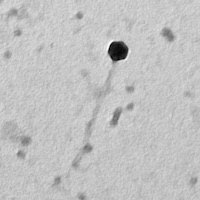
| Plaque Notes | William produces clear plaques the relatively round and small, ranging from approximately 1 mm to 3 mm in length. |
| Morphotype | Siphoviridae |
| Number of Genes | 83 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewWilliam_1 William_2 William_3 William_4 William_5 William_6 William_7 William_8 William_9 William_10 William_11 William_12 William_13 William_14 William_15 William_16 William_17 William_18 William_19 William_20 William_21 William_22 William_23 William_24 William_25 William_26 William_27 William_28 William_29 William_30 William_31 William_32 William_33 William_34 William_35 William_36 William_37 William_38 William_39 William_40 William_41 William_42 William_43 William_44 William_45 William_46 William_47 William_48 William_49 William_50 William_51 William_52 William_53 William_54 William_55 William_56 William_57 William_58 William_59 William_60 William_61 William_62 William_63 William_64 William_65 William_66 William_67 William_68 William_69 William_70 William_71 William_72 William_73 William_74 William_75 William_76 William_77 William_78 William_79 William_80 William_81 William_82 William_83 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MK801721 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 42 |
| Pitt Freezer Box Grid# | H12 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |