| Detailed Information for Phage Zarbodnamra | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Armando Braz |
| Year Found | 2017 |
| Location Found | Bethlehem, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.65659 N, 75.342957 W Map |
| Discovery Notes | Location: This soil sample was collected from my hometown of Bethlehem, PA just prior to the start of the semester. I wanted to see if I'd see any luck with some soil from over 300 miles away, just out of curiosity. The Soil Conditions: I retrieved this soil from my vegetable garden that had been used the previous summer. This soil is used annually to grow tomatoes, peppers, cucumbers, parsley, broccoli and other herbs/vegetables. Every year I use miracle grow nutrients in order to give the plants more of what they need to grow. On the day of soil collection (Jan 8, 2017), the sol was very dry and frozen. The soil appeared "deadened" in that the soil was a gray brown colour and was not very cohesive (sandy/loamy texture). There was a lot of plant detritus mixed in with the soil. Weather Conditions: At the time of collection, air temperature was -6 degrees celcius. 3 days prior there was a snow storm, with snow from that still present on the ground. Weather over the past 24 hours involved blustering winds, with freezing rain overnight. During the actual collection of the soil however, skies were clear. |
| Naming Notes | Note: the "(r)'s" are silent. Pronounced zah-bod-nom-ah. "We are going to die, and that makes us the lucky ones. Most people are never going to die because they are never going to be born. The potential people who could have been here in my place but who will in fact never see the light of day outnumber the sand grains of Arabia. Certainly those unborn ghosts include greater poets than Keats, scientists greater than Newton. We know this because the set of possible people allowed by our DNA so massively exceeds the set of actual people. In the teeth of these stupefying odds it is you and I, in our ordinariness, that are here. We privileged few, who won the lottery of birth against all odds, how dare we whine at our inevitable return to that prior state from which the vast majority have never stirred?" —Richard Dawkins |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 16, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 1504 |
| Genome length (bp) | 49729 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | TCGCCGGTGA |
| GC Content | 67.3% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CV |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
Click to ViewArchis Azula Barco Begonia BlingBling Blino Blueberry CaptainKirk2 CarolAnn CheeseSpider Delian Fairfaxidum Fenry Frokostdame Gambino Guacamole Hitter Jalammah JasperJr JuJu KappaFarmDelta Lysidious Malachai Melba Mellie MintFen MissRona Murp Obliviate PCoral7 Petra PhrostedPhlake Pipp PrincePatrick Samba Toast UmaThurman Utz Walrus William Wisp Wocket Zarbodnamra ZiggyZoo Zilla |
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
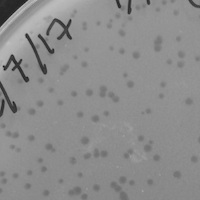
| Plaque Notes | My phage produced clear plaques that vary in diameter size, ranging from diameter = .5mm to diameter = 3mm. The plaques are for the most part entirely circular and form a well defined boundary between where cells underwent lysis, and where cells did not. |
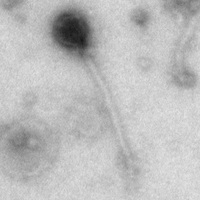
| Morphotype | Siphoviridae |
| Number of Genes | 84 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewZarbodnamra_1 Zarbodnamra_2 Zarbodnamra_3 Zarbodnamra_4 Zarbodnamra_5 Zarbodnamra_6 Zarbodnamra_7 Zarbodnamra_8 Zarbodnamra_9 Zarbodnamra_10 Zarbodnamra_11 Zarbodnamra_12 Zarbodnamra_13 Zarbodnamra_14 Zarbodnamra_15 Zarbodnamra_16 Zarbodnamra_17 Zarbodnamra_18 Zarbodnamra_19 Zarbodnamra_20 Zarbodnamra_21 Zarbodnamra_22 Zarbodnamra_23 Zarbodnamra_24 Zarbodnamra_25 Zarbodnamra_26 Zarbodnamra_27 Zarbodnamra_28 Zarbodnamra_29 Zarbodnamra_30 Zarbodnamra_31 Zarbodnamra_32 Zarbodnamra_33 Zarbodnamra_34 Zarbodnamra_35 Zarbodnamra_36 Zarbodnamra_37 Zarbodnamra_38 Zarbodnamra_39 Zarbodnamra_40 Zarbodnamra_41 Zarbodnamra_42 Zarbodnamra_43 Zarbodnamra_44 Zarbodnamra_45 Zarbodnamra_46 Zarbodnamra_47 Zarbodnamra_48 Zarbodnamra_49 Zarbodnamra_50 Zarbodnamra_51 Zarbodnamra_52 Zarbodnamra_53 Zarbodnamra_54 Zarbodnamra_55 Zarbodnamra_56 Zarbodnamra_57 Zarbodnamra_58 Zarbodnamra_59 Zarbodnamra_60 Zarbodnamra_61 Zarbodnamra_62 Zarbodnamra_63 Zarbodnamra_64 Zarbodnamra_65 Zarbodnamra_66 Zarbodnamra_67 Zarbodnamra_68 Zarbodnamra_69 Zarbodnamra_70 Zarbodnamra_71 Zarbodnamra_72 Zarbodnamra_73 Zarbodnamra_74 Zarbodnamra_75 Zarbodnamra_76 Zarbodnamra_77 Zarbodnamra_78 Zarbodnamra_79 Zarbodnamra_80 Zarbodnamra_81 Zarbodnamra_82 Zarbodnamra_83 Zarbodnamra_84 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH576969 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 56 |
| Pitt Freezer Box Grid# | F10 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |