| Detailed Information for Phage Affeca |
| Discovery Information |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Brett Rosenblatt |
| Year Found | 2017 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.443889 N, 79.955556 W Map |
| Discovery Notes | This phage was discovered of the William Pitt Union. The ground was frozen and there was snow around the digging site. The soil I took was initially frozen but, I let it sit in an insulated compartment of bag bag. |
| Naming Notes | This name came from a nickname of a friend I work with. I chose this name because I felt ut was unique name that has meaning to me. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Sep 29, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 1203 |
| Genome length (bp) | 59156 |
| Character of genome ends | Circularly Permuted |
| GC Content | 67.5% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | DE |
| Subcluster | DE1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
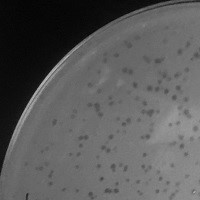
| Plaque Notes | This phage yields round, clear plaques. This phage does , however, appear to present itself in two different sized plaques. One, the size of the tip of a pencil point, and the other about 2x the size. |
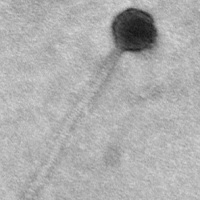
| Morphotype | Siphoviridae |
| Number of Genes | 87 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH976506 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
55 |
| Pitt Freezer Box Grid# |
B9 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |