
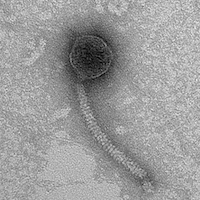
| Detailed Information for Phage Duplicity |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Alex Jennings |
| Year Found | 2014 |
| Location Found | Arkadelphia, AR USA |
| Finding Institution | Ouachita Baptist University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 34.126111 N, 93.052222 W Map |
| Naming Notes | When trying to isolate a single phage it looked to be like two phages in my plates and it wasn't until the 6th Titer Assay that we came to the conclusion that is ONE phage creating these plaques. So I have named it Duplicity because that means deceitful or two. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 15, 2020 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 1210 |
| Genome length (bp) | 42938 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 14 bases |
| Overhang Sequence | CCCGCCGCAATGGG |
| GC Content | 66.3% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | N |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Virginia Tech |
| Annotation Status | In GenBank |
| Morphotype | Siphoviridae |
| Number of Genes | 70 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | OL742561 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
5 x 10 ^13 |
| Pitt Freezer Box# |
8 |
| Pitt Freezer Box Grid# |
E4 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |