| Detailed Information for Phage Hanako | |
| Discovery Information | |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Lei Helton |
| Year Found | 2024 |
| Location Found | Concord, CA United States |
| Finding Institution | Lehigh University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 11°C |
| GPS Coordinates | 37.944677 N, 122.025127 E Map |
| Discovery Notes | The soil sample was collected from the surface of my backyard lawn after recent rain. The soil had a solid and muddy consistency and grainy texture; weather conditions at the time of collection were foggy. |
| Naming Notes | This phage is named after my older sister, Hanako, who has inspired and challenged me all our lives. Aspiring to both live up to her achievements and step out of her shadow pushes me to work harder for my goals every day- even pushing me to apply to this program. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jul 23, 2024 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 7578 |
| Genome length (bp) | 42595 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 13 bases |
| Overhang Sequence | CCCGCCGCCTTGG |
| GC Content | 66.1% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | N |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
Click to ViewAggie Andies BabeRuth Bosection6 Butters Carcharodon Charlie Chewbacca Cubone Duplicity EGUnicorn Fulbright Gex Hanako Impisi Jamie19 Journey Kevin1 Magsby Melville MichelleMyBell Nenae Panchino Parmesanjohn PhancyPhin Philonius Phloss Phrann Pipsqueaks Purgamenstris Raymond7 Rebel Redi Rubeelu Schnauzer Scitech ShrimpFriedEgg Shweta Silvafighter Silvy SkinnyPete Smurph Snekmaggedon Spinach SpongeBob Tapioca Tortoise12 Xeno Xerxes |
| Annotating Institution | Lehigh University |
| Annotation Status | In GenBank |
| Plaque Notes | After 48 hours, clear plaques ranging from 0.5-1 mm formed consistently. |
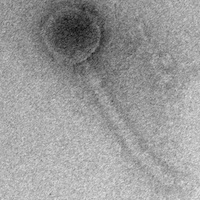
| Morphotype | Siphoviridae |
| Number of Genes | 69 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewHanako_1 Hanako_2 Hanako_3 Hanako_4 Hanako_5 Hanako_6 Hanako_7 Hanako_8 Hanako_9 Hanako_10 Hanako_11 Hanako_12 Hanako_13 Hanako_14 Hanako_15 Hanako_16 Hanako_17 Hanako_18 Hanako_19 Hanako_20 Hanako_21 Hanako_22 Hanako_23 Hanako_24 Hanako_25 Hanako_26 Hanako_27 Hanako_28 Hanako_29 Hanako_30 Hanako_31 Hanako_32 Hanako_33 Hanako_34 Hanako_35 Hanako_36 Hanako_37 Hanako_38 Hanako_39 Hanako_40 Hanako_41 Hanako_42 Hanako_43 Hanako_44 Hanako_45 Hanako_46 Hanako_47 Hanako_48 Hanako_49 Hanako_50 Hanako_51 Hanako_52 Hanako_53 Hanako_54 Hanako_55 Hanako_56 Hanako_57 Hanako_58 Hanako_59 Hanako_60 Hanako_61 Hanako_62 Hanako_63 Hanako_64 Hanako_65 Hanako_66 Hanako_67 Hanako_68 Hanako_69 |
| Submitted Minimal DNA Master File | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PQ723755 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| SEA Lysate Titer | 2.0 x 10^11 pfu/mL |
| Pitt Freezer Box# | 198 |
| Pitt Freezer Box Grid# | D6 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |