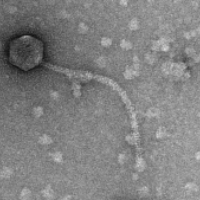
| Detailed Information for Phage ChickenTender | |
| Discovery Information | |
| Isolation Host | Gordonia rubripertincta NRRL B-16540 |
| Found By | Cooper Braun, Navin Kunakornvanich |
| Year Found | 2024 |
| Location Found | Spokane, WA United States of America |
| Finding Institution | Gonzaga University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 22°C |
| GPS Coordinates | 47.667817 N, 117.401007 W Map |
| Discovery Notes | Our sample was taken from the front of the oldest dorm in the middle of campus. The soil was pretty dry but maintained. We discovered it on a sunny but colder day in early fall, around the end of August. |
| Naming Notes | My partner and I just did the first thing that happened to pop into our heads so, chicken tender. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 13, 2025 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 3684 |
| Genome length (bp) | 45255 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGTAGGCTT |
| GC Content | 60.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CT |
| Subcluster | -- |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
Click to ViewAgatha AikoCarson Amok AndPeggy Axym Azira Balloona Bavilard BigChungus BillDoor Biskit Blondies Bubble Burnsey Button Buttrmlkdreams CanesSauce CaramelLatte Carsonalex CherryonLim ChickenTender ChocoMunchkin Cleo CocoaPuff Cozz Dre3 Elinal Eliott ElJefes Emalyn Feastonyeet Fribs8 Gibbous GiKK GoldHunter GTE2 Hexbug HippoPololi Horseradish Jamzy KayGee Lauer MAnor Margaret MaVan Mayweather McDazzle MScarn MunkgeeRoachy Nibbles Nina Nodigi Orla Pons PotPie PsychoKiller Quasar RADical RanchParmCat RedBaron RSchmailzl SheckWes SketchMex Socotra Sopespian Starburst SteamedHams SummitAcademy Survivors SweatNTears Tolls Troje Typhonomachy Vine Wolfwood Yakult Yarn Yucky Yummy Zareef |
| Annotating Institution | Gonzaga University |
| Annotation Status | Being Annotated (Expected completion by 2/1/2027) |
| Plaque Notes | The plaques tend to be on the smaller side and while still pretty round, they are slightly jagged. The plates and plaques are slightly turbid because the bacteria gives an orange hue. Along with the plaque clearings, theres a ring or halo of slight clearing around all of the plaques. |
| Morphotype | Siphoviridae |
| Number of Genes | 68 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewChickenTender_1 ChickenTender_2 ChickenTender_3 ChickenTender_4 ChickenTender_5 ChickenTender_6 ChickenTender_7 ChickenTender_8 ChickenTender_9 ChickenTender_10 ChickenTender_11 ChickenTender_12 ChickenTender_13 ChickenTender_14 ChickenTender_15 ChickenTender_16 ChickenTender_17 ChickenTender_18 ChickenTender_19 ChickenTender_20 ChickenTender_21 ChickenTender_22 ChickenTender_23 ChickenTender_24 ChickenTender_25 ChickenTender_26 ChickenTender_27 ChickenTender_28 ChickenTender_29 ChickenTender_30 ChickenTender_31 ChickenTender_32 ChickenTender_33 ChickenTender_34 ChickenTender_35 ChickenTender_36 ChickenTender_37 ChickenTender_38 ChickenTender_39 ChickenTender_40 ChickenTender_41 ChickenTender_42 ChickenTender_43 ChickenTender_44 ChickenTender_45 ChickenTender_46 ChickenTender_47 ChickenTender_48 ChickenTender_49 ChickenTender_50 ChickenTender_51 ChickenTender_52 ChickenTender_53 ChickenTender_54 ChickenTender_55 ChickenTender_56 ChickenTender_57 ChickenTender_58 ChickenTender_59 ChickenTender_60 ChickenTender_61 ChickenTender_62 ChickenTender_63 ChickenTender_64 ChickenTender_65 ChickenTender_66 ChickenTender_67 ChickenTender_68 |
| Publication Info | |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 192 |
| Pitt Freezer Box Grid# | C7 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |