| Detailed Information for Phage Niobe |
| Discovery Information |
| Isolation Host | Arthrobacter globiformis NRRL B-2880 |
| Found By | Erica Sellers |
| Year Found | 2020 |
| Location Found | New Haven, CT United States |
| Finding Institution | Southern Connecticut State University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 41.33349 N, 72.945666 W Map |
| Discovery Notes | Found at the edge of a garden near a building. Garden contained plants such as pachysandra, rhododendron, and some kind of ivy. The area had no disturbed soil indicating no recent animal activity, and was largely shaded by vegetation, getting little sunlight. The weather had ranged from 32.8°C-16.1°C in the week before collection. There had been devastating thunderstorms the week before collection.
Depth: 4cm
Temperature when collected: 18.3°C |
| Naming Notes | Named after the queen from Greek mythology. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 7, 2021 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 1729 |
| Genome length (bp) | 43602 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CGAAGGGGCAT |
| GC Content | 66.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | AZ |
| Subcluster | AZ1 |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Southern Connecticut State University |
| Annotation Status | In GenBank |
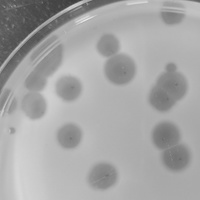
| Plaque Notes | Average of 8 mm in diameter, circular. Temperature morphology, clear inner circle surrounded by a cloudy outer circle. |
| Morphotype | Siphoviridae |
| Number of Genes | 70 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MZ820087 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
7.5e10 |
| Pitt Freezer Box# |
129 |
| Pitt Freezer Box Grid# |
F7 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |
