| Detailed Information for Phage KilKor |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Kiley Banks and Korrin Taylor |
| Year Found | 2018 |
| Location Found | Longview, TX USA |
| Finding Institution | LeTourneau University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 32.4655 N, 94.727 W Map |
| Discovery Notes | Excavation of the soil took place right below a tree in a flower bed outside of Glaske Center. We had to move flowers out of the way and dig past a layer of mulch to remove soil. We dug to a depth of 2 inches before taking the sample. We filled the collection tube to 3.5 ml with soil. Soil was a light brown and slightly sandy, but had good moisture. The soil was permeated by threadlike roots and contained two dense chunks of soil. |
| Naming Notes | The name comes from adding the first three letters from both of the founders' first names together. Kil(ey) + Kor(rin) = KilKor. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 11, 2019 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 1275 |
| Genome length (bp) | 48916 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 13 bases |
| Overhang Sequence | CCTGCCGCCCCGA |
| GC Content | 67.2% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | P |
| Subcluster | P1 |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | The Lysogeny of KilKor was the lysogenic reproductive cycle. |
| Other Cluster Members |
|
| Annotating Institution | LeTourneau University |
| Annotation Status | In GenBank |
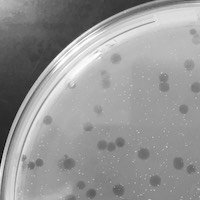
| Plaque Notes | The plaques are roughly 1-2.2mm in diameter. They are circular and clear in the center of the plaque, although they get slightly cloudy along the edge. |
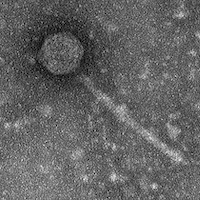
| Morphotype | Siphoviridae |
| Number of Genes | 79 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MN892486 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
85 |
| Pitt Freezer Box Grid# |
C9 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |