
| Detailed Information for Phage Newt |
| Discovery Information |
| Isolation Host | Gordonia terrae 3612 |
| Former names | Sami |
| Found By | Kanchan Paudel |
| Year Found | 2018 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.4434 N, 79.9538 W Map |
| Discovery Notes | Found in flower bed under approximately 4 inches of mulch. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 15, 2019 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina Sequencing |
| Approximate Shotgun Coverage | 598 |
| Genome length (bp) | 73414 |
| Character of genome ends | 3' Sticky Overhang |
| Direct Terminal Repeat Length | 151 bp |
| GC Content | 59.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | CS |
| Subcluster | CS3 |
| Cluster Life Cycle | Lytic |
| Lysogeny Notes | Hypothesized clear plaques with halos are indicative of a lytic phage
No further lysogen testing completed. |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
| Plaque Notes | Small (approximately 1.5 mm), circular plaques of roughly uniform size. Slight evidence of halos on edges of plaques. Center of the plaques are clear, indicative of a lytic phage. |
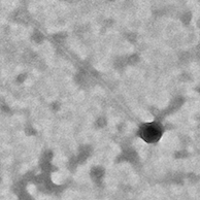
| Morphotype | Siphoviridae |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MK919481 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
1.0 x 10^10 pfu/mL |
| Pitt Freezer Box# |
90 |
| Pitt Freezer Box Grid# |
F12 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |