| Detailed Information for Phage Bjanes7 | |
| Discovery Information | |
| Isolation Host | Gordonia terrae 3612 |
| Found By | Linn Bjanes |
| Year Found | 2016 |
| Location Found | Pittsburgh, PA USA |
| Finding Institution | University of Pittsburgh |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 40.445 N, 79.953333 W Map |
| Discovery Notes | Identified area with sprouting new plants in a flower bed near the Cathedral of Learning. There was a fresh layer of snow on top of the dirt so I brushed away the snow and dug a few inches underneath the top soil. I dug roughly 3 inches into the earth about 2 inches from the nearest plant Collected 1 spoonful of dirt (the ground was frozen) Placed the soil sample into the plastic bag and sealed the bag with the zip closures Recorded the GPS coordinates using the compass app (see below) After roughly 5 minutes of transport, the soil sample was put into a conical tube for enrichment Additional Notes: GPS coordinates of the soil collection location: 40˚26’42” N 79˚57’12” W Weather conditions at the time of the collection: strong winds, overcast with snow showers, 34 degrees F Weather conditions in the last 24 hours: frequent snow showers, gusting winds and mostly cloudy; high 20s during the day and low 10s during the night Soil conditions: soil was moist and mixed in with grass, mulch and bits of snow and ice. |
| Naming Notes | Last name: Bjanes and 7 because it was in sample #7 that the phage was discovered. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jun 30, 2016 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 2827 |
| Genome length (bp) | 46042 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | TACCAGGGGGA |
| GC Content | 66.6% |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | CZ |
| Subcluster | CZ2 |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | A theory about the lysogeny is that because several morphologies present themselves (small and large/turbid) the phage uses both the lytic cycle and the lysogenic cycle. This is not supported by an immunity assay as of now but may be done later. |
| Other Cluster Members |
Click to ViewBunnybear (None) Lton (None) Ohgeesy (None) Pepperoni (None) Agueybana (CZ1) AlumE (CZ1) Antonio (CZ1) BatStarr (CZ1) Bialota (CZ1) Bosnia (CZ1) BoyNamedSue (CZ1) Eudoria (CZ1) Eviarto (CZ1) Herod (CZ1) Hugley (CZ1) Hyzer (CZ1) Kita (CZ1) Madavieve (CZ1) Madeline (CZ1) Manasvini (CZ1) Maridalia (CZ1) Neobush (CZ1) Nymphadora (CZ1) Polly (CZ1) Suscepit (CZ1) Tayonia (CZ1) ThankyouJordi (CZ1) TimTam (CZ1) Trumpet (CZ1) WelcomeAyanna (CZ1) Zameen (CZ1) Zirinka (CZ1) Attis (CZ2) Bjanes7 (CZ2) Clap (CZ2) Cynthia (CZ2) Ebert (CZ2) EdnaMode (CZ2) FroggyToad (CZ2) GemG (CZ2) Gizermo (CZ2) Haley23 (CZ2) Jalleen (CZ2) JCole (CZ2) Lamberg (CZ2) LordFarquaad (CZ2) Matteo (CZ2) Mocha12 (CZ2) Nettuno (CZ2) Pickett (CZ2) Sahara (CZ2) Savage (CZ2) SoilAssassin (CZ2) Sproutie (CZ2) TuertoX (CZ2) Vasanti (CZ2) Whiteclaw (CZ2) Yeet412 (CZ2) BaxterFox (CZ3) PantheRoc (CZ3) Yeezy (CZ3) Adora (CZ4) Annalisa (CZ4) Beenie (CZ4) Clark (CZ4) DobbysSock (CZ4) Dolores (CZ4) Dorito (CZ4) Easley (CZ4) Hortense (CZ4) Howe (CZ4) Invectra (CZ4) Mcklovin (CZ4) MichaelScott (CZ4) Oregano (CZ4) PhriskyACE (CZ4) Powerball (CZ4) Samman98 (CZ4) Sekhmet (CZ4) Shlim410 (CZ4) Suerte (CZ4) Thimann (CZ4) Tuti (CZ4) Twinkle (CZ4) WinkNick (CZ4) Denise (CZ5) Faith5x5 (CZ6) Moosehead (CZ6) OneDirection (CZ6) Sidious (CZ7) RavenCo17 (CZ8) |
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
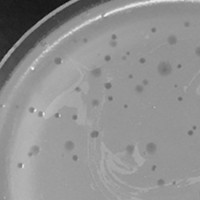
| Plaque Notes | The plaque morphologies are both small, clear plaques and large, turbid plaques. The large ones have haloes, hazy outer edges and a tiny, clear dot in the center. |
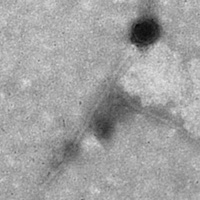
| Morphotype | Siphoviridae |
| Number of Genes | 72 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
Click to ViewBjanes7_1 Bjanes7_2 Bjanes7_3 Bjanes7_4 Bjanes7_5 Bjanes7_6 Bjanes7_7 Bjanes7_8 Bjanes7_9 Bjanes7_10 Bjanes7_11 Bjanes7_12 Bjanes7_13 Bjanes7_14 Bjanes7_15 Bjanes7_16 Bjanes7_17 Bjanes7_18 Bjanes7_19 Bjanes7_20 Bjanes7_21 Bjanes7_22 Bjanes7_23 Bjanes7_24 Bjanes7_25 Bjanes7_26 Bjanes7_27 Bjanes7_28 Bjanes7_29 Bjanes7_30 Bjanes7_31 Bjanes7_32 Bjanes7_33 Bjanes7_34 Bjanes7_35 Bjanes7_36 Bjanes7_37 Bjanes7_38 Bjanes7_39 Bjanes7_40 Bjanes7_41 Bjanes7_42 Bjanes7_43 Bjanes7_44 Bjanes7_45 Bjanes7_46 Bjanes7_47 Bjanes7_48 Bjanes7_49 Bjanes7_50 Bjanes7_51 Bjanes7_52 Bjanes7_53 Bjanes7_54 Bjanes7_55 Bjanes7_56 Bjanes7_57 Bjanes7_58 Bjanes7_59 Bjanes7_60 Bjanes7_61 Bjanes7_62 Bjanes7_63 Bjanes7_64 Bjanes7_65 Bjanes7_66 Bjanes7_67 Bjanes7_68 Bjanes7_69 Bjanes7_70 Bjanes7_71 Bjanes7_72 |
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info | |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MG099941 |
| Refseq Number | None yet |
| Archiving Info | |
| Archiving status | Archived |
| Pitt Freezer Box# | 3 |
| Pitt Freezer Box Grid# | C5 |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |