
| Detailed Information for Phage Brewer |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Zoey Brewer adopted by Malia Harris |
| Year Found | 2025 |
| Location Found | Helena, MT United States |
| Finding Institution | Montana Technological University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 37°C |
| GPS Coordinates | 46.60022 N, 112.05404 W Map |
| Discovery Notes | Discovered June 2nd, 2025, from indoor garden starter by Zoey Brewer. Part of Jared Hunt's class at C.R. Anderson Middle School in Helena, MT. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 6, 2026 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina NextSeq 1000 |
| Read Type | Single-end reads |
| Read Length | 100 bp |
| Approximate Shotgun Coverage | 858 |
| Genome length (bp) | 73945 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 9 bases |
| Overhang Sequence | CGCTTGTCA |
| GC Content | 62.9% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | E |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | Brewer lysogen was resistant to infection by supernatant of Brewer phage along with Brewer phage lysate. Brewer lysogen and Brewer lysate gave off phage DNA on the Wild type plate and killed Mycobacterium smegmatis bacteria. |
| Other Cluster Members |
|
| Annotating Institution | Montana Technological University |
| Annotation Status | Being Annotated (Expected completion by 5/1/2026) |
| Plaque Notes | Smaller plaque sizes with halo ring around plaques. |
| Number of Genes | 136 |
| Number of tRNAs | 2 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | No |
| GenBank Accession | None yet |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| Virtual Digest Picture | Download |
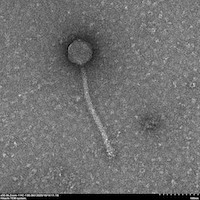
| EM Picture | Download |