| Detailed Information for Phage Inca |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Courtney Davies |
| Year Found | 2015 |
| Location Found | Auckland, New Zealand |
| Finding Institution | Massey University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 36.671139 S, 174.662436 E Map |
| Discovery Notes | Soil sample collected 27th August 2015 at 9.57am.
The ambient temperature was 13°C. No wind and a light drizzle of rain at time of collection. A small hole was dug and the soil collected 2cm deep underneath a rotting silage bale. The silage had not been grazed by cattle since April 2015 and had started to decompose. The site was fenced off with an electric fence above and was 5m east of a concreted driveway on a flat rural/lifestyle property in Dairy Flat. |
| Naming Notes | Inca is the name of my purebred Ayrshire stud. I chose the same name as it represents my cows whom are very special to me (the initial name stemmed from the successful Inca Empire). Inca can also be roughly translated to "ruler" or "lord". So not only does Inca Phage have a fascinating name, it is also an intriguing phage full of potential! |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Nov 29, 2015 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 2299 |
| Genome length (bp) | 74070 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 9 bases |
| Overhang Sequence | CGCTTGTCA |
| GC Content | 62.8% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | E |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Massey University |
| Annotation Status | In GenBank |
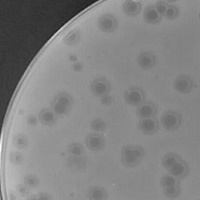
| Plaque Notes | The morphology was very interesting with this plaque. Initially there were lytic plaques present between 24 and 72 hours of incubation, however after 10 days of continued incubation at 37°C these changed and it was clear that the once lytic plaque now had a turbid ring around it and a further lytic ring beyond. From there on there was no change. Plaques were approximately 2mm initially and then developed into 5mm plaques. |
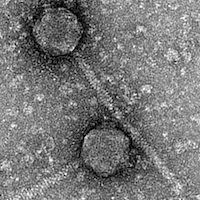
| Morphotype | Siphoviridae |
| Number of Genes | 145 |
| Number of tRNAs | 2 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH576956 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
2.0 x 10^9 pfu/mL |
| Date of SEA Lysate Titering |
Sep 18, 2015 |
| Pitt Freezer Box# |
19 |
| Pitt Freezer Box Grid# |
C11 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |