| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Gretchen Walch |
| Year Found | 2012 |
| Location Found | Alexandria, KY USA |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 38.93909 N, 84.37412 W Map |
| Discovery Notes | Soil was captured on the edge of a creek, 50 cm (about)from the closest water edge; water was moving but congested at the nearest point to the water. The air temperature at the time was about 26.1 degrees Celsius. The area surrounding the sample location was muddy and interspersed with grass, plat roots, and an earth worm was captured, but releases upon collecting the sample. It was within 30 cm of larger creek vegetation/weeds and flowers. The location was approximately 115.7 cm under a wood bridge thus receiving shade throughout the day at differing times and intensity. The closest tree was within approximately 4-5 meters of the location. The man made things in the area included a bed of concrete within 115.5 cm the nearest road was approximately 16 meters away. The location was aptly located in a sort of valley with hills facing East and West with the creek itself running South to North. It is known to have frequent flooding reaching 2-3 meters of the creek edge (thus the soil sample location is within the flood zone). Finally, it is known that the creek could have septic run-off from the surrounding houses running along the road. The creek itself stems from a pond 1.5 miles South of the sample location and runs from the pond North (1.5 miles) to the sample capture location. The creek is exposed to the run-off of 5 West facing houses (2 of which are on the hill of the valley) and 6 East facing houses all of which are on the East facing hill. (houses are within 400 yards and are in rural spacing.) |
| Naming Notes | The name of Simpliphy came from the idea that phages are in essence simple as in small and not complex in structure. However, despite this they have many possible applications that can be very helpful. For example, wound healing. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | May 23, 2013 |
| Sequencing Facility | Virginia Commonwealth University Nucleic Acids Research Facilities |
| Shotgun Sequencing Method | 454 |
| Sequencer Used | 454 Genome Sequencer FLX |
| Approximate Shotgun Coverage | 112 |
| Genome length (bp) | 75366 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 9 bases |
| Overhang Sequence | CGCTTGTCA |
| GC Content | 63.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | E |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Western Kentucky University |
| Annotation Status | In GenBank |
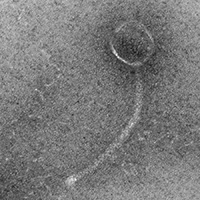
| Morphotype | Siphoviridae |
| Number of Genes | 145 |
| Number of tRNAs | 2 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH536827 |
| Refseq Number | None yet |
| Published in Paper | Pope, Bowman, Russell, et al 2015, eLIFE: Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
43 |
| Pitt Freezer Box Grid# |
16 |
| Available Files |
| EM Picture | Download |
| GenBank File for Phamerator | Download |