| Detailed Information for Phage Xandras |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Ashley Gledhill |
| Year Found | 2023 |
| Location Found | Beaver Dam, KY United States |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 30°C |
| GPS Coordinates | 37.46371 N, 86.8124 W Map |
| Discovery Notes | Dirt sample was collected from within a chicken coop, no visible grass growing in dirt, very compact and a bit moist. |
| Naming Notes | I hope this phage will one day be used in phage therapy, so I chose the name Xandras because "Xandra" means "defender of the people |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 16, 2024 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina Sequencing |
| Approximate Shotgun Coverage | 1039 |
| Genome length (bp) | 75179 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 9 bases |
| Overhang Sequence | CGCTTGTCA |
| GC Content | 62.9% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | E |
| Subcluster | -- |
| Cluster Life Cycle | Temperate |
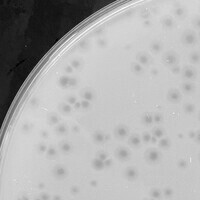
| Lysogeny Notes | A lysogen was isolated, purified, and grown in a broth, and then dilutions of Xandras were plated on a lawn of the lysogen, confirming immunity |
| Other Cluster Members |
|
| Annotating Institution | Western Kentucky University |
| Annotation Status | In GenBank |
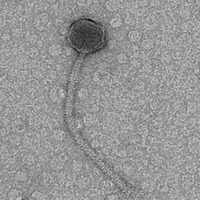
| Morphotype | Siphoviridae |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PP750962 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |