| Detailed Information for Phage Craff |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Gregory Maas |
| Year Found | 2016 |
| Location Found | Evansville, IN USA |
| Finding Institution | University of Evansville |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | Not entered |
| GPS Coordinates | 37.971129 N, 87.531267 W Map |
| Discovery Notes | found sample in flowerbed in sesquicentennial oval at the University of Evansville |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 21, 2016 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 307 |
| Genome length (bp) | 69263 |
| Character of genome ends | Circularly Permuted |
| GC Content | 66.4% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | B |
| Subcluster | B1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | University of Evansville |
| Annotation Status | In GenBank |
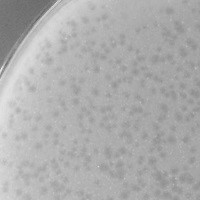
| Plaque Notes | medium, cloudy |
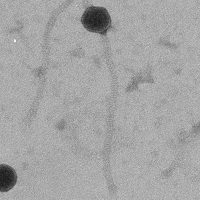
| Morphotype | Siphoviridae |
| Number of Genes | 103 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MH399773 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
3.66x10^10 |
| Date of SEA Lysate Titering |
Nov 10, 2016 |
| Pitt Freezer Box# |
31 |
| Pitt Freezer Box Grid# |
A5 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |