Mycobacterium phage LasagnaCat
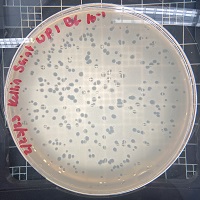
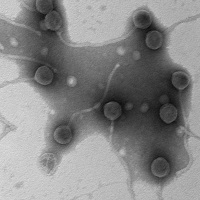
Know something about this phage that we don't? Modify its data.
Recently Added Phages
WileyE Ditch Greece Sway SupergirlRecently Modified Phages
Mudpuppy Kalizoi Karkharias Pitbull MellowYellowRecently Finished Phages
Torres (K1) Inyanga (A1) Vagabond (O) Quantum (BM) ThayneTheZag (AY)
