| Detailed Information for Phage KLucky39 |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Kyler Haskell |
| Year Found | 2009 |
| Location Found | Provo, UT USA |
| Finding Institution | Brigham Young University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 40.2464 N, 111.648822 W Map |
| Discovery Notes | Date: 15 Sep 2009
Temp: 24°C
Site: BYU Campus by Widstoe Building
Sample: Topsoil <1 in., wet mulch in bark. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Mar 17, 2011 |
| Sequencing Facility | Joint Genomics Institute |
| Shotgun Sequencing Method | Sanger |
| Sequencer Used | Sanger ABI 3730 |
| Read Type | Paired-end reads |
| Genome length (bp) | 68138 |
| Character of genome ends | Circularly Permuted |
| GC Content | 66.5% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | B |
| Subcluster | B1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
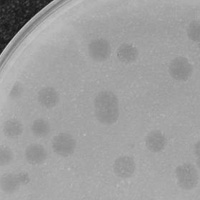
| Plaque Notes | Plaques are small, 1mm, and lysogenic with cloudy concentric circles around the 1mm clear spot. |
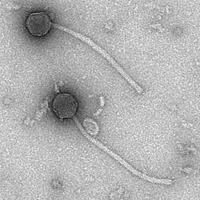
| Morphotype | Siphoviridae |
| Number of Genes | 100 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | JF704099 |
| Refseq Number | None yet |
| Published in Paper | Pope, Bowman, Russell, et al 2015, eLIFE: Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity |
| Archiving Info |
| Archiving status |
Archived |
| SEA Designator |
2009BRYOklucky39KH |
| SEA Lysate Titer |
1e5 |
| Date of SEA Lysate Titering |
Oct 13, 2009 |
| Pitt Freezer Box# |
11 |
| Pitt Freezer Box Grid# |
72 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| Virtual Digest Picture | Download |
| EM Picture | Download |