| Detailed Information for Phage Bonray |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | John Brackett |
| Year Found | 2017 |
| Location Found | Dahlonega, GA USA |
| Finding Institution | University of North Georgia |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 34.533056 N, 83.985 W Map |
| Discovery Notes | Found in well-established Urban garden bed with flowers and tree. There had been no recent rain and the temperature was 72 degrees Fahrenheit.The soil was fine and slightly damp. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Dec 21, 2017 |
| Sequencing Facility | Pittsburgh Bacteriophage Institute |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 493 |
| Genome length (bp) | 157364 |
| Character of genome ends | Circularly Permuted |
| GC Content | 64.7% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | C |
| Subcluster | C1 |
| Cluster Life Cycle | Lytic |
| Other Cluster Members |
|
| Annotating Institution | University of Pittsburgh |
| Annotation Status | In GenBank |
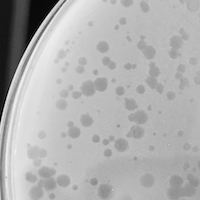
| Plaque Notes | Bonray, produces plaques that are consistent in all aspects except size. The varying in sizes may be due to rapid mutation of the phage. The largest plaques are roughly 1.7mm, the medium ones about .63mm and the smallest plaques are .19mm. The large size of some plaques and the smooth edges indicates my phage may be a rapid lysis mutant. The plaques are clear and there is almost no turbidity. |
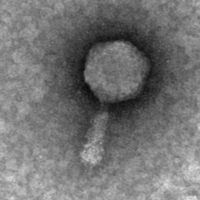
| Morphotype | Myoviridae |
| Number of Genes | 236 |
| Number of tRNAs | 33 |
| Number of tmRNAs | 1 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MK112533 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
5x10e9 |
| Pitt Freezer Box# |
74 |
| Pitt Freezer Box Grid# |
G3 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |