
| Detailed Information for Phage Groupthink |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Colleen Jackson |
| Year Found | 2015 |
| Location Found | Bowling Green, KY USA |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 36.9854 N, 86.45 W Map |
| Discovery Notes | My soil sample was gathered from rocky and fertilized ground between Western Kentucky University's two greenhouses. The soil had fertilizer from what was used within the greenhouse and thus likely had abnormally high levels of nutrients in comparison to other close locations. In addition, the location did not have much vegetation with the exception of crabgrass. The area also had smaller amount of sunlight due to the presence of the buildings and tree nearby. Temperature at time of sample was 37.5 degrees. |
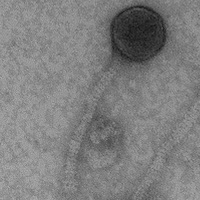
| Naming Notes | The name is derived from the behavior of the phage. The Phage capsids were very attracted to each other during the electron microscopy, hence the name Groupthink. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Mar 1, 2016 |
| Shotgun Sequencing Method | Illumina |
| Approximate Shotgun Coverage | 8373 |
| Genome length (bp) | 50574 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGGTGGTAA |
| GC Content | 64.0% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | A |
| Subcluster | A3 |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | The phage crated very turbid plaques at 37 degrees for 24 hours. The was development of clear plaque mutants at a high concentration of phage on the plate. |
| Other Cluster Members |
|
| Annotating Institution | Western Kentucky University |
| Annotation Status | In GenBank |
| Plaque Notes | This was a very turbid plaque which could not be accurately depicted by pictures. This phage developed clear plague mutants after the population became "webby" on the plates. In addition the plagues sizes uniformly decreased as the plate became more populated. |
| Morphotype | Siphoviridae |
| Number of Genes | 89 |
| Number of tRNAs | 3 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MK967399 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Designator |
2015WEKUgroupthinkCSJ |
| SEA Lysate Titer |
4.4 x 10^9 pfu/ml |
| Date of SEA Lysate Titering |
Oct 29, 2015 |
| Pitt Freezer Box# |
27 |
| Pitt Freezer Box Grid# |
E6 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |