| Detailed Information for Phage Fastidio |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Hyunbi H, Ethan W, Payson D, Matt J |
| Year Found | 2023 |
| Location Found | Provo, UT United States |
| Finding Institution | Brigham Young University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| Isolation Temperature | 29°C |
| GPS Coordinates | 40.214582 N, 111.618295 W Map |
| Discovery Notes | Collected from soil in Utah County. |
| Naming Notes | I had to do a process called 10 plate a couple times in order to create a sufficiently large high-titer lysate. This annoyed me a little bit because I didn't understand why it wasn't working. Therefore I named it "fastidio" which is annoyance in spanish. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Apr 24, 2024 |
| Sequencing Facility | CD Genomics |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 3593 |
| Genome length (bp) | 55839 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 10 bases |
| Overhang Sequence | CGGACGGCGC |
| GC Content | 61.6% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | F |
| Subcluster | F1 |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Brigham Young University |
| Annotation Status | In GenBank |
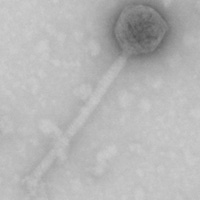
| Morphotype | Siphoviridae |
| Number of Genes | 108 |
| Number of tRNAs | 0 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | PQ244011 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Not in Pitt Archives |
| Available Files |
| EM Picture | Download |
| GenBank File for Phamerator | Download |
