| Detailed Information for Phage BGlluviae |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Trong Phung |
| Year Found | 2016 |
| Location Found | Bowling Green, KY USA |
| Finding Institution | Centre College |
| Program | Kentucky Biomedical Infrastructure Network Small Genomes Discovery Program |
| From enriched soil sample? | Yes |
| Isolation Temperature | Not entered |
| GPS Coordinates | 36.956944 N, 86.463889 W Map |
| Discovery Notes | Soil collected on 1/3/16 at 11:44 am at a depth of 6 centimeters. The temperature was about 51 degrees farenheit, 11 degrees celcius. The soil was moist and a grey-black color. The soil is collected about 10 feet away from a storm drain in a semi-urban area and 25 feet away from the nearest street. The location of collection was also surrounded by vegetation, mostly grasses and weeds. Little foot traffic passes here. Unsloped in a storm basin. |
| Naming Notes | The soil is from Bowling Green, Kentucky and from a storm drain. So I gave the initial BG, and then lluviae is derived from the Spanish word "lluvia" meaning rain. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Mar 20, 2019 |
| Sequencing Facility | NC State Genomic Sciences Laboratory |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 235 |
| Genome length (bp) | 59308 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CTCGTAGGCAT |
| GC Content | 66.5% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | K |
| Subcluster | K1 |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Centre College |
| Annotation Status | In GenBank |
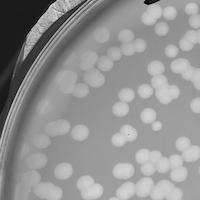
| Plaque Notes | The plaques are circular, clear and usually about 2.5 mm in size. They have somewhat a fast period of full growth, usually less than 15 hours. |
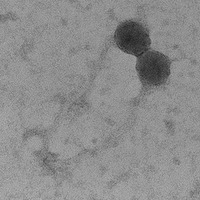
| Morphotype | Siphoviridae |
| Number of Genes | 95 |
| Number of tRNAs | 1 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MN908692 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
1.8x10^10 pfu/mL |
| Date of SEA Lysate Titering |
Feb 15, 2016 |
| Pitt Freezer Box# |
27 |
| Pitt Freezer Box Grid# |
G2 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |