| Detailed Information for Phage Inky |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Keith Miller |
| Year Found | 2016 |
| Location Found | Louisville, KY USA |
| Finding Institution | Spalding University |
| Program | Kentucky Biomedical Infrastructure Network Small Genomes Discovery Program |
| From enriched soil sample? | Yes |
| Isolation Temperature | 12°C |
| GPS Coordinates | 38.0225 N, 85.715556 W Map |
| Discovery Notes | Brooks, KY
30 yards into a wooded area,hill top.
many leaves on the ground
soil sample taken at 5 cm depth |
| Naming Notes | Favorite ghost from PacMan |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Nov 30, 2018 |
| Sequencing Facility | NC State Genomic Sciences Laboratory |
| Shotgun Sequencing Method | Illumina |
| Sequencer Used | Illumina MiSeq |
| Read Type | Single-end reads |
| Read Length | 150 bp |
| Approximate Shotgun Coverage | 260 |
| Genome length (bp) | 59708 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CTCGTAGGCAT |
| GC Content | 66.5% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | K |
| Subcluster | K1 |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | Spalding University |
| Annotation Status | In GenBank |
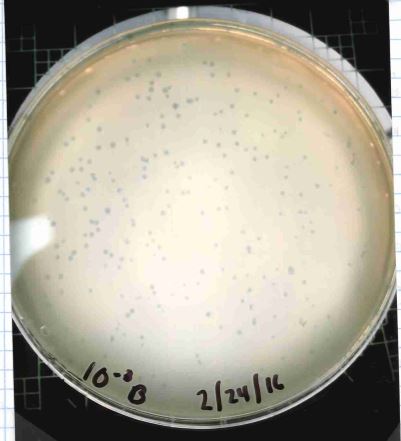
| Plaque Notes | clear plaques, 1.5 mm diameter after 24 hour incubation at 37C |
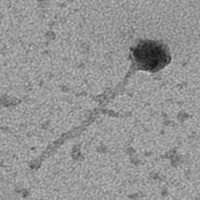
| Morphotype | Siphoviridae |
| Number of Genes | 95 |
| Number of tRNAs | 1 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Minimal DNA Master File | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MN369746 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| Pitt Freezer Box# |
28 |
| Pitt Freezer Box Grid# |
B9 |
| Available Files |
| Plaque Picture | Download |
| EM Picture | Download |
| GenBank File for Phamerator | Download |