| Detailed Information for Phage Juliette |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Laura Johnson |
| Year Found | 2012 |
| Location Found | Holland, MI USA |
| Finding Institution | Hope College |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| Isolation Temperature | 37°C |
| GPS Coordinates | 42.78 N, 86.1 W Map |
| Naming Notes | This phage was named after Juliette from Romeo and Juliette as the literary Juliette tends to leave a wake of death behind her which is appropriate for a virus. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Aug 8, 2019 |
| Sequencing Facility | Hope College |
| Shotgun Sequencing Method | Illumina |
| Approximate Shotgun Coverage | 37 |
| Genome length (bp) | 58071 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CTCGCGGCCAT |
| GC Content | 67.5% |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | K |
| Subcluster | K4 |
| Cluster Life Cycle | Temperate |
| Other Cluster Members |
|
| Annotating Institution | University of Mary |
| Annotation Status | In GenBank |
| Morphotype | Siphoviridae |
| Number of Genes | 96 |
| Number of tRNAs | 1 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MW601218 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Designator |
2012HOPEjulietteLEJ |
| SEA Lysate Titer |
1.75 * 10^11 |
| Date of SEA Lysate Titering |
Oct 23, 2012 |
| Pitt Freezer Box# |
36 |
| Pitt Freezer Box Grid# |
48 |
| Available Files |
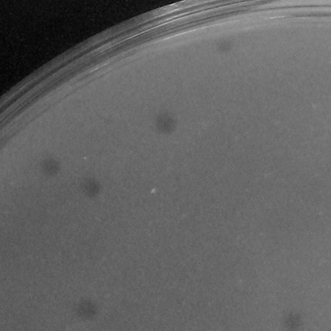
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
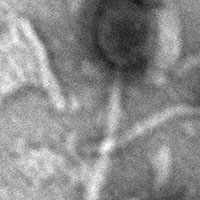
| EM Picture | Download |
| GenBank File for Phamerator | Download |