| Detailed Information for Phage Jeckyll |
| Discovery Information |
| Isolation Host | Mycobacterium smegmatis mc²155 |
| Found By | Taylor Senay |
| Year Found | 2015 |
| Location Found | Elizabethtown, KY USA |
| Finding Institution | Western Kentucky University |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Unknown |
| Isolation Temperature | 37°C |
| GPS Coordinates | 37.725833 N, 85.862778 W Map |
| Discovery Notes | Soil sample was obtained from an area about a meter from the walking/biking trail with light foot traffic. The soil itself was dry, taken from about 5cm under a fallen tree limb. The surrounding area was wooded, with no buildings or concrete in sight. Sample was taken from an area approximately 30m from a small standing creek. Air temperature was approximately 30 degrees Celsius. |
| Naming Notes | This phage exhibited particularly aggressive clear plaque mutants. This brought to mind the story of Dr. Jekyll and Mr. Hyde. Since the name Jekyll was already taken, I opted for a slight alteration in spelling.
See Sequencing Notes below for information about the sequencing of one of these clear plaque mutants. |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Apr 5, 2016 |
| Shotgun Sequencing Method | Illumina |
| Approximate Shotgun Coverage | 350 |
| Genome length (bp) | 59708 |
| Character of genome ends | 3' Sticky Overhang |
| Overhang Length | 11 bases |
| Overhang Sequence | CTCGTAGGCAT |
| GC Content | 66.6% |
| Sequencing Notes | A clear plaque mutant of Jeckyll was isolated and sequenced. It had two differences from the wild type Jeckyll.
G instead of A at position 34912.
G instead of A at position 41250. |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | K |
| Subcluster | K1 |
| Cluster Life Cycle | Temperate |
| Lysogeny Notes | Temperate phage |
| Other Cluster Members |
|
| Annotating Institution | Unknown or unassigned |
| Annotation Status | In GenBank |
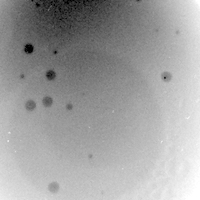
| Plaque Notes | Turbid plaques, typically 3-4mm in diameter, with small clear centers. Incubated at 37˚C for 48 hours to produce plaques this size |
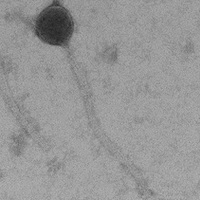
| Morphotype | Siphoviridae |
| Number of Genes | 96 |
| Number of tRNAs | 1 |
| Number of tmRNAs | 0 |
| Has been Phamerated? | Yes |
| Gene List |
|
| Submitted Annotation in DNA Master Format | Download |
| Submitted Minimal DNA Master File | Download |
| Author List | Download |
| Annotation Cover Sheet | Download |
| Publication Info |
| Uploaded to GenBank? | Yes |
| GenBank Accession | MF140412 |
| Refseq Number | None yet |
| Archiving Info |
| Archiving status |
Archived |
| SEA Lysate Titer |
8.4x10^8 |
| Pitt Freezer Box# |
27 |
| Pitt Freezer Box Grid# |
E11 |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |
| Final DNAMaster File | Download |
| GenBank File for Phamerator | Download |